Brain Tissue Segmentation
Expectation Maximization with a Gaussian Mixture Model
Brain tissue segmentation using an Expectation Maximization (EM) algorithm for Gaussian Mixture Models (GMM)
Authors: Frederik Hartmann, Xavier Beltran Urbano
Code: Github
Report: Github
This project was carried out within the scope of the Medical Image Segmentation course taught by Prof. Dr. Xavier Lladó. You can reach a more mathematical report here.
October 2023
Dataset
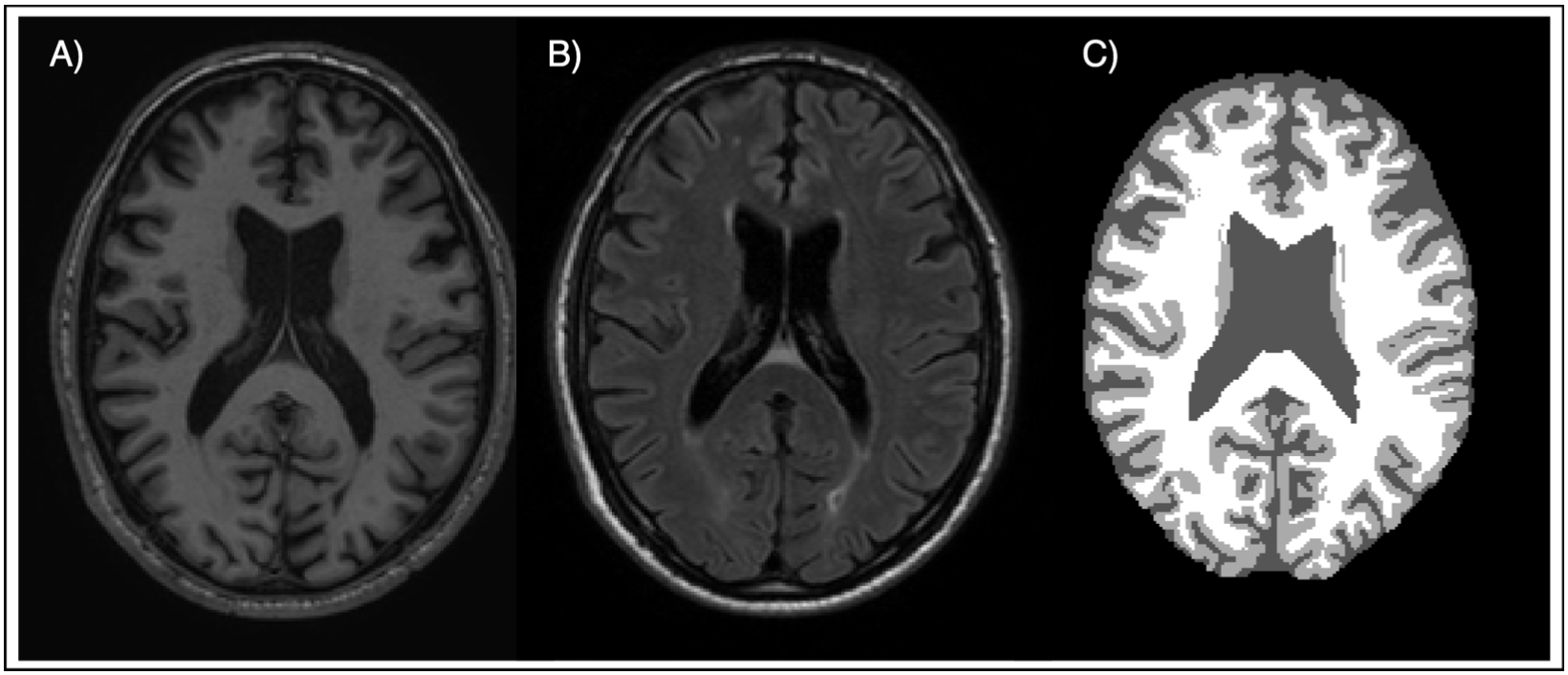
The segmentation process was conducted using a multimodal dataset comprising 5 MRI images, each with the following modalities:
- T1-weighted (T1)
- FLAIR-weighted (FLAIR)
- Ground Truth (GT)
Methodology
Our approach utilizes the EM algorithm in two primary steps:
- Expectation Step: Here, we assign a membership weight to each voxel, indicating the likelihood of it belonging to a particular tissue cluster.
- Maximization Step: In this step, we update the parameters of our Gaussian models — means, covariances, and mixture weights — based on the current membership assignments.
The model initializes with either random parameters or using KMeans clustering for a more informed starting point. The iterative process of expectation and maximization continues until convergence is achieved, indicated by the log-likelihood reaching a plateau.
Results
The algorithm’s effectiveness was quantitatively evaluated using the Dice Score (DSC), a metric that computes the similarity between predicted and ground truth segmentations. Tables 1 and 2 present the algorithm’s performance metrics alongside comparisons with other segmentation tools.
Table 1: Evaluation results of the Expectation Maximization Algorithm on the test set
| Initialization | Modalities | GM [DSC] | WM [DSC] | CSF [DSC] | Iterations | Time [s] |
|---|---|---|---|---|---|---|
| KMeans | T1 | 0.8457 ± 0.0436 | 0.8015 ± 0.0300 | 0.8120 ± 0.0251 | 116 ± 4 | 24.36 ± 1.28 |
| KMeans | T1+FLAIR | 0.8457 ± 0.0436 | 0.8017 ± 0.0304 | 0.8129 ± 0.0249 | 110 ± 17 | 51.48 ± 8.10 |
| Random | T1 | 0.8025 ± 0.0819 | 0.7284 ± 0.1383 | 0.7860 ± 0.0670 | 346 ± 108 | 65.87 ± 18.42 |
| Random | T1+FLAIR | 0.8468 ± 0.0450 | 0.7978 ± 0.0233 | 0.8082 ± 0.0278 | 333 ± 117 | 147.71 ± 51.16 |
Table 2: Evaluation results of KMeans and SPM
| Method | Initialization | Modalities | GM [DSC] | WM [DSC] | CSF [DSC] | Time [s] |
|---|---|---|---|---|---|---|
| K-means | K-means ++ | T1 | 0.8222 ± 0.0356 | 0.7464 ± 0.0338 | 0.8542 ± 0.0078 | 3.57 ± 0.12 |
| K-means | K-means ++ | T1+FLAIR | 0.8243 ± 0.0357 | 0.7500 ± 0.0315 | 0.8560 ± 0.0071 | 3.61 ± 0.19 |
| SPM | - | T1 | 0.75 ± 0.03 | 0.81 ± 0.03 | 0.76 ± 0.02 | - |
Conclusion
In conclusion, the application of the Expectation Maximization algorithm with Gaussian Mixture Models has proven to be an effective strategy for the segmentation of brain tissues in MRI images. Our methodology not only aligns with the stringent requirements of medical image processing but also showcases the potential to facilitate early diagnosis and treatment planning for neurodegenerative diseases.